
Health Insurance Cost Prediction and Regression
Insurance-Cost-Prediction Health Insurance Cost Prediction and Regression
Read More






Published on May 26, 2017 by Samer on ML
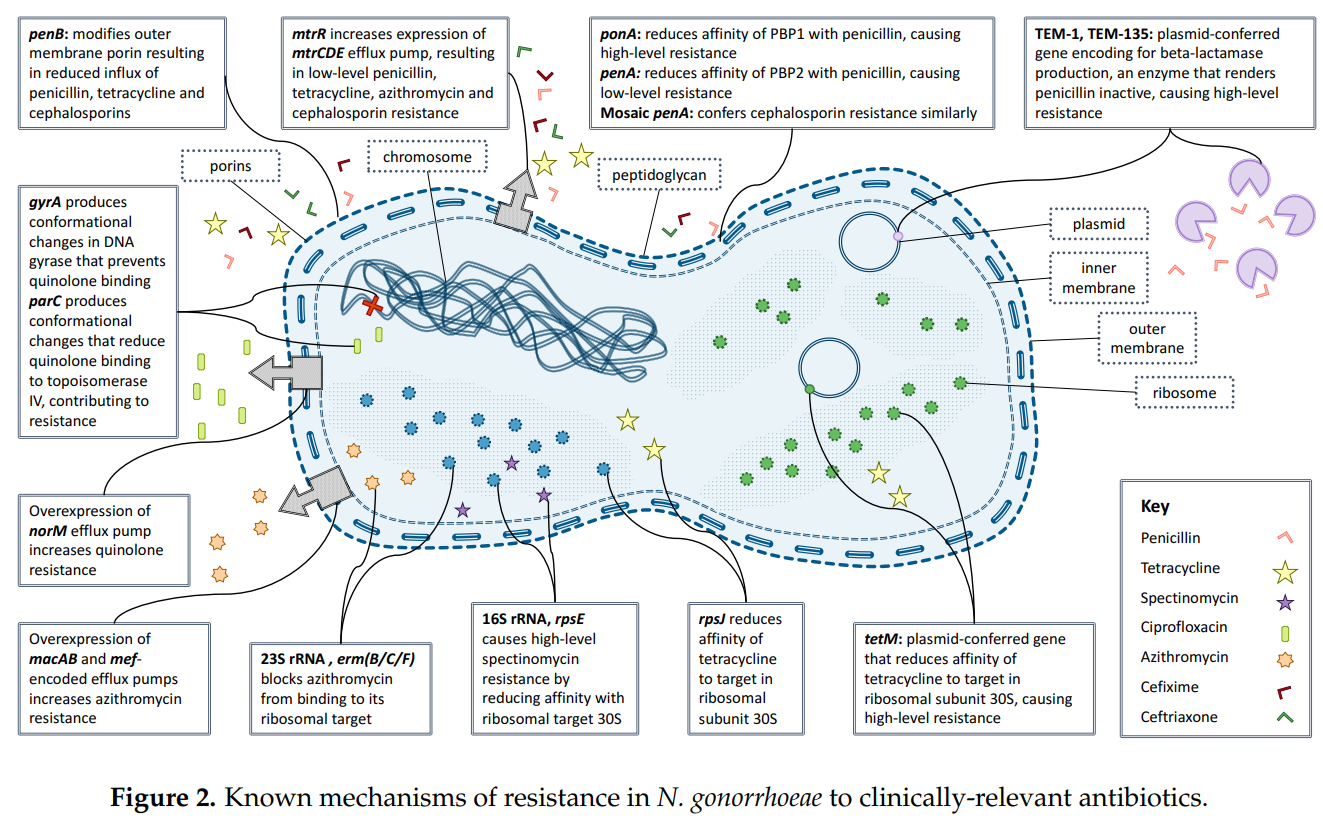
Antibiotic-DNA-resistant-sequence In this project, you’ll have a machine learning model for predicting antibiotic resistance in bacteria. I will be focussing on a species called Neisseria gonorrhoeae, the bacteria which cause gonorrhoea. Gonorrhoea is the second most common sexually transmitted infection (STI) in Europe, after chlamydia. Rates of gonorrhoea infection are on the rise, with a 26% increase reported from 2017-2018 in the UK. Many people who are infected (especially women) experience no symptoms, helping the disease to spread. If the infection is left untreated, it can lead to infertility in women, and can occasionally spread to other parts of the body such as your joints, heart valves, brain or spinal cord. Resistance of these bacteria to antibiotics is rising over time, making infections hard to treat. Below, you can see rates of resistance to different antibiotics.
Images and references are from the paper: Epidemiological Trends of Antibiotic Resistant Gonorrhoea in the United Kingdom Lilith K. Whittles 1, Peter J. White 1,2,3,4, John Paul 5,6 and Xavier Didelot 1, Published: 13 July 2018 Can be accessed on that link: https://www.mdpi.com/2079-6382/7/3/60
In the past, patients were treated with an antibiotic called ciprofloxaxcin. Doctors had to stop using this antibiotic because resistance to the drug became too common, causing treatments of infections to fail. Until very recently, the recommended treatment was two drugs - ceftriaxone and azithromycin. Azithromycin was removed from recommendations because of concern over rising resistance to the antibiotic. In February 2018, the first ever reported case of resistance to treatment with ceftriaxone and azithromycin, as well as resistance to the last-resort treatment spectinomycin, was reported. Currently in the UK, patients are only treated with ceftriaxone. In this notebook, you will look at machine learning algorithms for predicting resistance to three drugs (Azithromycin – Cefixime – Ciprofloxacin)
For this work, we will be using scikit learn, pandas numpy matplotlib seaborn
For this Notebook, we have genome sequence and antibiotic resistance data gathered from publicly available sources microreact (project N_gonorrhoeae).You can have an interactive view of the data can be accessed Neisseria gonorrhoea antibiotic resistance (microreact.org) For this analysis, I’m using unitigs, are available on NCBI on this link Nucleotide BLAST: Search nucleotide databases using a nucleotide query (nih.gov) and filtered by DBGWAS project, unitigs are short stretches of DNA shared by a subset of the strains in our study. Unitigs are an efficient but flexible way of representing DNA variation in bacteria. You can read more about unitigs and DBGWAS project at this link: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1007758
The full dataset consists of 584,362 unitigs, which takes a long time to train models on, so for this exercise we will be using a set that has been filtered for unitigs statistically associated with resistanceusing built-in filter in the DBGWAS project. We have data on resistance to several different antibiotics. We call the different resistance patterns ‘phenotypes’, which is a general term for traits that an organism has.
We want to be able to quickly build models using the same process for different traits, as we will go through three different antibiotics, so we can make a function to get the data ready for model building that takes the phenotype as input and returns the relevant information. Methods 1-data prepare The information we want is the presence/absence pattern of the unitigs across each sample (X) and how resistant each sample is to an antibiotic (pheno). So, we first build a function for preparing our training and testing data and filter it by dropping samples that don’t have a value for our chosen resistan
Then read our unitig data and only keep rows with a resistance measure prepare our data for predicting antibiotic resistance create an array for storing performance metrics have a look at the data to be sure and have a n idea of our used data
2-Model training We will start will building some basic models for azithromycin resistance. This resistance pattern can mostly be explained by a single mutation, so is likely to be impacted by the amount of noise each method incorporates. When we train a machine learning model, we want to know how well the model will work on new data. we can look at this is to use ‘cross validation’. so, we perform K-fold cross validation, where we split the data into 5 parts and we build 5 models, each with a part of the data hidden from the model and used to measure its accuracy.
Then we perform grid search to identify best hyper-parameters, predict resistance in test set which will be built on calling all the samples with a predicted value less than or equal to 0.5 as sensitive to the antibiotic, and samples with predicted value >0.5 resistant to the antibiotic.
We can go through fitting different models. First, we will try an elastic net logistic regression, support vector machine, XGBoost and finally, a random forest.
Results part
-compare accuracies and times from the different predictors by using curves and plotting
Look into what your model has learned, and whether this fits with our existing knowledge of antibiotic resistance -looking at SVM feature importance -looking at random forest feature importance -plotting and print the unitigs, allows knowing what genes related to input one into this search algorithm: https://card.mcmaster.ca/analyze/blast to see if it comes from a known resistance mechanism. and you will need to choose the BLASTN option for the query to be processed correctly, to confirm our results and to know if our two algorithms learned roughly the same thing? Does what they learned make sense?
Examine how much we’d benefit from collecting more samples to be sure Should we collect more samples? Or not Here, we will construct a learning curve to see how much performance improves as we include more of our sample set, and whether performance gains have levelled off, suggesting that collecting more samples won’t help, or whether it’s still improving. • Then we predict resistance for another antibiotic (Azithromycin – Cefixime – Ciprofloxacin) in order.
more projects Click on image to visit my website 👇
Antibiotic-DNA-resistant-sequence
Want to run this project in your computer Follow these Steps Clone or Download
Open the terminal/CMD in project directory
Then create virtual environment using this command:
py -m venv env
Activate virtual environment using:
env\Scripts\activate
Install all the requirements using:
scikit learn, pandas, numpy, matplotlib and seaborn
It will take some time to download till that take a sip of coffee ☕
After successful download of all above requirements, run the app using:
python library

Insurance-Cost-Prediction Health Insurance Cost Prediction and Regression
Read More
Data is the lifeblood of many organizations and the quality of that data can greatly impact the decisions made from it. The process of cleaning and transforming data is known...
Read More
Disease-diagnosing-Prediction using machine learning different alghorithms to predict disease. Currently this app is deployed in a Tkinter App, and provides 3 different high accurate clinical diagnosis based on 3 different...
Read More